Advanced BioTech Solutions for Healthcare
Department of Pharmaceutical Sciences, Bouve College of Health Sciences
Biotechnology research
Microfluidic devices, with their nano-liter reaction volumes and parallel sample processing, are ideal for total chemical and bioassay analysis. These capabilities enable ultra-high throughput screening and efficient use of limited samples and reagents, driving innovation in research and diagnostics.
read more...
Our lab is focused on developing novel Bio-MEMS approaches to advance point of care diagnostics, cell culture and drug screening and delivery methods. We have developed Lab-on-a-Chip (LOC) devices that integrate several laboratory functions such as real time monitoring of target clinically relevant analyte, proteomics, genomics, cell-cell interactions as well as cell secretion and surface monitoring of single cells on a micro-chip. The advantages of the microfluidic based LOC developed by our lab include, the pico and nano-liter volumes, elimination of cross-contamination, the fast and efficient mixing of the reagents and gases and the ability to manipulate and analyze cells at a very high-throughput.
Our droplet based bio-assay platforms:
Biomedical research
Advancements in cell-based therapy are revolutionizing immune response modulation, offering new potential for treating a range of conditions. Our microfluidics technology accelerates research and innovation in this cutting-edge field, providing precise control for cell-based applications
read more...
A promising approach to facilitate immune activation is the adoption of tissue engineering technology to generate three-dimensional (3D) constructs for cell function, cell-cell interaction studies, drug screening as well as cells and drugs delivery. We have developed a droplet microfluidic based technology to generate and monitor effects of therapeutic regimen on biomimetic micro-tumors tissue-chip. The microfluidic device is capable of generation and in-situ analysis of the micro-tumors for different applications in a high-throughput manner. It is equipped with a continuous perfusion module for continuous drug treatment and renewal of media and nutrients for the experimental duration to mimic in vivo conditions.
Furthermore, our laboratory is investigating a localized tissue engineered approach using 3D biocompatible materials to construct bio-matrixes loaded with vaccine and cells to enhance the immunologic milieu in patients. These constructs may serve as “lymphoid-like” constructs engineered to be implanted in vivo to deliver long-term immunity and specifically target cells to induce necessary mechanisms.
Cell migration and interactions in our bioengineered lymphoid-like constructs
read more...
The research program in our lab is focused on two general categories: 1. Development, evaluation and delivery of new therapeutic strategies and 2. Development of novel diagnostic and detection tools. In particular, we are working on a novel approach to develop and deliver cell-based therapy for modulating the immune response in cancer via bioengineered “lymphoid-like” constructs while our Lab-on-a-Chip (LOC) devices applied towards both analysis and manipulation of immunological system in cancer patients to overcome the disease.
Furthermore, we are applying our novel biotech methods to develop robust and portable device for detection of time-varying concentrations in real-time, of soluble molecules such as dosed insulin in T1D patients and phenotypic-based antimicrobial susceptibility testing (AST). Successful implementation of these research activities will represent a transformative advance in the field of diagnostics, therapy, biosensors, pharmacokinetic analysis and immunoassay development in microfluidic format.
Latest Research
List of IP and Patents
- INV-14119 Microfluidic device and Method for Analysis of Tumor Microenvironments: US-2017-0199173-A1
- INV-16003 Microdroplet Based Bioassay Platform: US 2020/0116709-A1
- INV-17084 Parallel Microfluidic Device for High Throughput Single Cell Assays in Microdroplets: US 20210308681 A1
- INV-17085 Live Single-Cell Bioassay In Micro-Droplets: US-2018-0313844-A1
- INV-19067 Microfluidic Chip for Single Cell Pairing: US 20220276227 A1
- INV-19076 Microfluidic Device for High Throughput Screening of Tumor Cell Adhesion and Motility: US 20220212192 A1
- INV-20077 Single Cell Isolation and Processing System with Reversible Well Shape: US 20210260576 A1
- INV-22098 A microfluidic platform integrated with on-demand droplet trapping and releasing for single cell omics analysis: WO 2023/215608
- INV-23014 Epigenetic Analysis After Single Cell Interactions Using Microfluidics: Pending
Single cell functional multi-omic analysis
Given the phenotypic and functional diversity of immune and cancer cell subsets, and the disparate timescale of immune reactions (early vs. delayed and transient vs. stable responses) it is essential to assess immune cell signaling and its target cell responses at single-cell level and more importantly in dynamic fashion to reveal the extent of heterogeneity. Our lab has developed h a microchip based method to evaluate cells and cellular interactions that can provide both high throughput and high content information bridging the gap between traditional assay plate in vitro methods and animal/human models.

Tumor on-a-chip
Konry lab has developed an in-situ droplet microfluidic based technology to generate and monitor effects of therapeutic regimen on biomimetic micro-tumor tissues. The microfluidic device is capable of generation and in-situ analysis of the immunogenic micro-tumors for different applications in a high-throughput manner. It is equipped with a continuous perfusion module for continuous drug treatment and renewal of media and nutrients for the experimental duration to mimic in vivo conditions. Since 2016, our published in Lab on a chip journal breast tissue-chip model was numerously cited and serves as a basis for our current tissue-chip studies.
We have also extended our microfluidic approach to establish Diffuse Large B cell lymphoma (DLBCL). Diffuse large B cell lymphoma (DLBCL), the most common subtype of Non-Hodgkin lymphoma, exhibits pathologic heterogeneity and a dynamic immunogenic tumor microenvironment (TME). However, the lack of preclinical in vitro models of DLBCL TME hinders optimal therapeutic screening. Thus we have focused on the development of an integrated platform for high-throughput generation of immunogenic DLBCL models in a novel hydrogel combination of alginate and puramatrix, which promoted cell adhesion and aggregation. We demonstrated this system in dynamic analysis of cellular interaction, proliferation and therapeutic efficacy via spatiotemporal monitoring and secretome profiling. This work was recently accepted to Journal of Controlled Release (2019).


Single cell droplet selection
Most cellular analysis methods use a top-down approach, examining proteomics or genomics directly and destroying the cell, which prevents correlating genomics with functional assays in the same cell. We present a bottom-up approach using single-cell tools. Our technology allows functional phenotyping by observing cell cytotoxicity and probing the underlying biology. We developed a droplet microfluidic device that traps and selectively releases droplets using microvalves. Each droplet contains natural killer (NK) cells and tumor cells for real-time monitoring of their interactions. We can release droplets with specific functional phenotypes, such as NK cells that rapidly kill tumor cells. This device enables the study of cell interactions, real-time monitoring, and cell recovery for single-cell analysis like RNA sequencing.
Another platform tangential to our droplet selection device we have developed is designed for high-throughput sorting and analysis of individual cells or particles within microscale droplets. It features an integrated system comprising microfluidic channels, fluorescence detection, and electrodes, allowing for precise control over sorting and collection. With this platform we seek to develop the capabilities to sort based on encapsulated functional cell activity.

Featured Articles
2021

Acoustofluidic Droplet Sorter Based on Single Phase Focused Transducers
2022

Droplet microfluidics for functional temporal analysis and cell recovery on demand using microvalves: application in immunotherapies for cancer
2023

Cellular immunity analysis by a modular acoustofluidic platform: CIAMAP
2024

Characterizing influence of rCHOP treatment on diffuse large B-cell lymphoma microenvironment through in vitro microfluidic spheroid model
The Konry lab developed a novel high throughput function to omic droplet microfluidic technology that can predict the clinical outcome in diffuse large B-cell lymphoma patients. The team deployed a microfluidic chip that can accurately replicate and predict the effect of chemotherapies such as Rituximab and CHOP combination on the ability of the NK cells to kill cancer cells. This achievement works to advance accurate immune profiling and function to omic analysis for discovery of new therapies and persolized medicine. Hyperlink to article
People
Research Team
Dr. Tania Konry, Principal Investigator
Jose Estevam, PhD Student
Michael Finocchiaro, PhD student
Christina Sharkey, PhD Student
Harry Akligoh, PhD Student
Abhishek Srihari Rampelli, PhD Student
Judene Thomas, PhD Student
Rachel White, PhD Student
Sujata Chalise, PhD Student
Masters students
Former Postdocs
Dr. Noa Cohen
Dr. Yantao Fan
Dr. Evangelia Hondroulis
Dr. Wenjing Kang
Dr. Alex Lim
Dr. Saheli Sarkar
Dr. Stefano Ugolini
Dr. Yichao Yang
Dr. Agustin De Ganzó
Former Grad Students
Ilana Berger Fridman
Pooja Sabhachandani
Matt Sullivan
Former Master and Undergrad Students
Anna Muranova, Jane Healy, Jake Saccoccio, Aditi Dave, Akhila Konda, Arushi Sharma, Chinmayee Sharma, Sneha Yadav, Shreya Nelson Singh, Yuntong Wang, Zhi Lin Shen, Millicent Gabriel, Awdhoot Godbole, Radhika Kulkarni, Ria Rajesh Bedi, Xinyi Shao, Suchitra Ramesh, Supriya Nagarajan, Viren BhatiaMichael Gray, Amey Gaikwad, Seamus McKenney, Kristy Fang, Sai Mynampati, Abhishek Chiyyeadu, Sayalee Potdar, Sneha Pawar, Rucha Adhav, Chanchal Rathi, Himali Shroff, Chaitra Belgur, Dipen Parande, Vinny Motwani, Abhinav Gupta, Sneha Varghese, Dishant Patel, Harnil Shah, Micah AmdurClark, Gillian Snyder, Allen Guo, Deborah Effiong, Sharon Berman
Latest news
The Konry lab was awarded the Sanofi Idea award for 2023 and 2024
The Konry lab is working on applying their functionomics approach to immunotheraputic applications in collaboration with Sanofi.
The Konry lab was awarded the
NSF CBET award (2023 – 2027)
The Konry lab was awarded the AHA grant for 2024 in collaboration with Boston Children’s hospital and Harvard
The Konry lab has been working in collaboration with the Bezzerides lab at Boston Children’s hospital since 2022 to functionally characterize modified cardiac cells on our microfluidic platform. Paving the way for functional and therapeutic applications.
Publication in Science Advances
Cellular immunity analysis by a modular acoustofluidic platform: CIAMAP (2024)
The Konry lab has published an article describing a modular acoustofluidic platform to systematically sort droplets based on NK92 and K562 co-encapsulation. The modularity of the platform enables of a droplet loading array for image analysis and rapid off loading for downstream analysis. Droplets populations are separated using a novel acoustofluidic pairing module integrated into the device. This platform will enable high-purity, easy-of-operation, real-time, and parallel cellular immunity analysis. More information can be found through the google scholar link. By Tania Konry, Ruoyu Zhong, Matthew Sullivan, and co-workers.
Associate Professor Dr. Tania Konry has been appointed as the new Assistant Dean for Research in the SOPPS effective 2022.
Dr. Tania Konry will serve as a member of the executive leadership team of the SOPPS and be responsible for providing administrative oversight and coordination for all activities pertaining to the research enterprise, and for enhancing and marketing the research reputation of the SOPPS at Bouvé College of Health Sciences and beyond by developing and implementing strategic initiatives to promote and sustain a culture of scholarship, research productivity and collegiality within the SOPPS and across the College and University.
The Konry Lab congratulates our PI on her new position!
New collaboration in 3D modelling applied to Multiple Myeloma malignancies between Dr. Irene Ghobrial – Dana Farber Cancer Institute and Dr. Tania Konry – NEU.
The project will be focused on the development of a microfluidics-based system applied to Multiple Myeloma malignancies in order to help improve the understanding of the immune tumor microenvironment.
The Konry Lab is looking forward to work with Dr. Ghobrial and her team at the Dana Farber Cancer Institute.
Publication in Small
Acoustofluidic Droplet Sorter Based on Single Phase Focused Transducers (Small 46/2021)
The cover image depicts the design of an acoustofluidic droplet sorter based on single phase focused transducers. In the droplet sorter, the droplets sequentially flow through a tapered channel where an optical fiber pair detects fluorescence signals and then, according to the fluorescence information, pulse acoustic signals are generated to trigger the single-phase focused transducers. The droplet is manipulated by standing surface acoustic waves and guided to the collection outlet if identified as a cell-laden droplet. Otherwise, the droplet maintains its route to flow into the waste outlet. More information can be found in article number 2103848 by Tania Konry, Tony Jun Huang, and co-workers.
Dr. Konry was awarded with CTSI Tufts and RADX/NIH Phase 0 grants for her PCR free RAPID testing technology for COVID-19 diagnostics
Collaboration in immunotherapy between Konry Lab NEU and Dr.R.Romee Dana Farber
Drs.Konry and Romee were awarder with a grant from DFCI/NEU joined program in cancer drug development (2019-2021). The grant is focused on applying novel microfluidic technology to develop memory NK cell based therapy.
Women in Microfluidics & BioMEMS
Dr.Konry was appointed to Women in Microfluidics & BioMEMS list.
Casis tissue NASA
Konry lab was awarded a collaborative NASA tissue grant to study the effect of the extreme environment of space on immune cell interactions.
Dr.Konry was awarded with 2017 Tufts Clinical and Translational Science Institute (CTSI) Pilot Award
The project will focus on determining antibody treatment sensitivity in B cell lymphoma by novel microfluidics-based NK cell immunogenicity platform developed by Dr.Konry’s group (Collaboration with Dr. Andrew Evens , Tufts Medical Center).
Pooja Sabhachandani will present at SLAS 2017 conference our work on novel droplet based platform for biomimetic tumor microenvironment studies and conduct the SLAS 2017 – Podium Presentation Webinar
Dr.Konry was nominated for SLAS Innovation Award 2017.
Dr.Konry was selected for funding by CIMIT’s NIH POC Award
The project is focused on developing a device for rapid urinary tract infection diagnosis and antibiotic susceptibility testing.
Our new findings in dynamic analysis of immune and cancer cell interactions: a single cell lab on a chip perspective
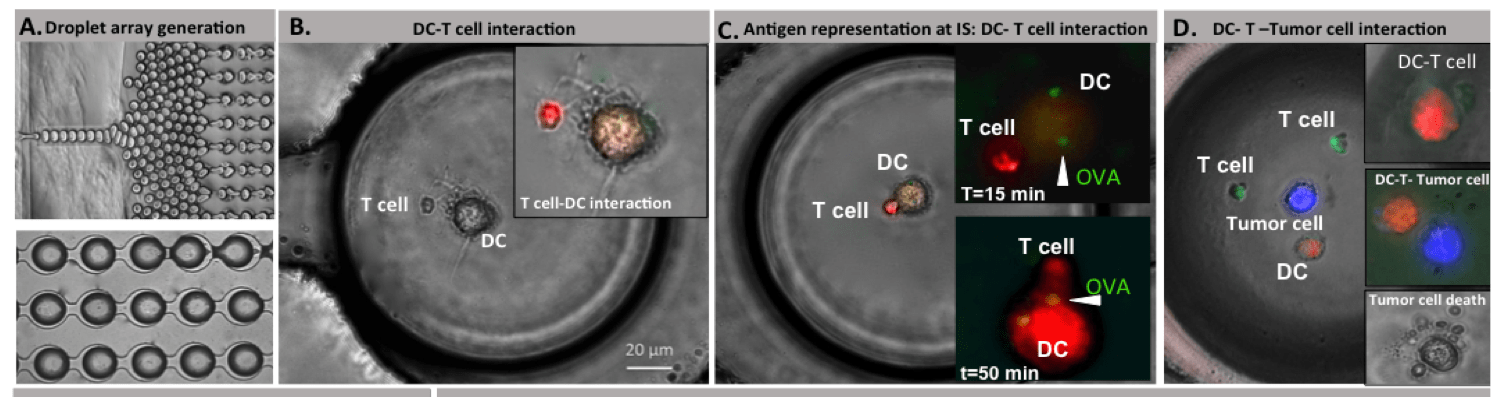
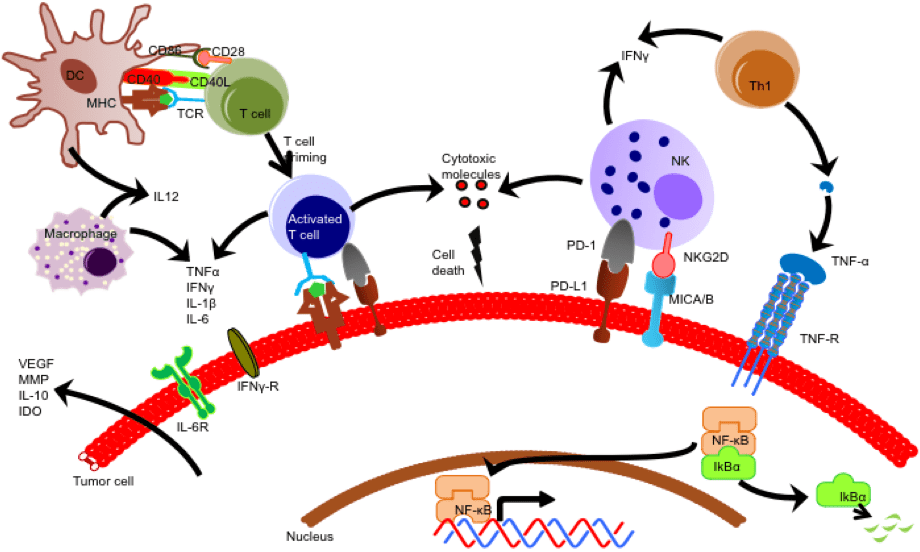
We investigated the dynamics of live cell anti-tumor immune responses at the single cell level in a microfluidic platform. The integrated droplet array allowed improved control over heterotypic cell pairing and interactions, which allowed us to observe significant cell motility, morphological changes, and complex formation over an extended duration. We evaluated immune cell priming by Ag-loaded DCs and the subsequent functional outcome in the con- text of multiple myeloma cells. Our results demonstrate substantial heterogeneity in priming interactions between DC and T cells, both in basal and activated cells. Effector T cells depicted time-varying cytotoxicity following transient, short or long stable contacts. Serial interactions by T cells were observed both in upstream (DC-based) and downstream (target-based) interactions. Our future aims include determining the molecular mechanisms underlying the phenotypic heterogeneity in T cell responses in droplets, and integrated live cell analysis of immune cell activation and effector functions.
Our novel droplet‐merging platform has been applied for comparative functional analysis of M1 and M2 macrophages for Forsyth institute
In our recent paper published in Biotechnology and Bioengineering we presented a simple and effective method for the co-encapsulation of polarized M1 and M2 macrophages with Escherichia coli (E. coli) by passive merging in an integrated droplet generation, merging, and docking platform. This approach facilitated live cell profiling of effector immune functions in situ and quantitative functional analysis of macrophage heterogeneity.
Our recent publication in Lab on a chip:
Phenotypic drug profiling in droplet microfluidics for better targeting of drug-resistant tumors
Acquired drug resistance is a key factor in the failure of chemotherapy. Due to intratumoral heterogeneity, cancer cells depict variations in intracellular drug uptake and efflux at the single cell level, which may not be detectable in bulk assays. In this study we present a droplet microfluidics-based approach to assess the dynamics of drug uptake, efflux and cytotoxicity in drug-sensitive and drug-resistant breast cancer cells. An integrated droplet generation and docking microarray was utilized to encapsulate single cells as well as homotypic cell aggregates. Drug-sensitive cells showed greater death in the presence or absence of Doxorubicin (Dox) compared to the drug-resistant cells. We observed heterogeneous Dox uptake in individual drug-sensitive cells while the drug-resistant cells showed uniformly low uptake and retention. Dox-resistant cells were classified into distinct subsets based on their efflux properties. Cells that showed longer retention of extracellular reagents also demonstrated maximal death. We further observed homotypic fusion of both cell types in droplets, which resulted in increased cell survival in the presence of high doses of Dox. Our results establish the applicability of this microfluidic platform for quantitative drug screening in single cells and multicellular interactions.
Dr. Konry was awarded with Schumacher Faculty Award 2015, presented to one faculty member early in their Northeastern career to recognize significant academic achievement for work done at Northeastern University
Dr. Konry was awarded with competitive grant from DFCI/NU Joint Program in Cancer Drug Development in collaboration with Prof. Suzanne Gaudet (DF/HMS).
Dr. Konry was announced as Phase 1 finalist of the Follow that Cell Challenge (NIH)
Dr. Konry was awarded with BD Biosciences research grant
Dr. Konry was awarded with FY16 TIER 1 Award for development of Highly Sensitive, Field Based, Lab on a Chip (LOC) for Serological Studies of Ebola Virus
Contact us
Tania (Tali) Konry, Ph.D. Associate Professor Department of Pharmaceutical Sciences Northeastern University 140 The Fenway, R 441, Lab 446 Boston, MA 02115 Tel: 617.373.3224
